FCU_guanylylCyclase.cellml
About this model
This is a Functional cell Unit (FCU) of the action of the soluble guanylyl cyclase (sGC) enzyme in the vascular smooth muscle cell, leading to contraction.
- INPUTS:
- Nitric oxide (NO)
- OUTPUTS:
- Stress generated by crossbridge contraction
- REACTIONS:
- Re1sGC: Transition of sGC from the basal state (EB) into an intermediate state (E6c) through NO binding
- Re2sGC: Natural decay of E6c to active E5c state
- Re3sGC: NO-dependent conversion of E6c into E5c
- Re4sGC: Conversion of E5c to EB through NO dissociation
- ReDNO: Consumption of NO by scavengers
- Re1a and 1b: Production of cyclic guanosine monophosphate (cGMP) from guanosine triphosphate (GTP) by E5c
- Re2a and 2b: Degradation of cGMP to GMP by phosphodiesterase-5 (PDE)
- Re12m: Phosphorylation of myosin light chain (M) to Mp
- Re34m: Binding of actin (A) to Mpto form cross bridge (AMp)
- Re56m: Cross bridge formation by phosphorylation of a latch bridge (AM)
- Re78m: Detachment of latch bridge into A and M
Model status
The current CellML implementation runs in OpenCOR.
Model overview
This model is based on existing kinetic model by Yang et. al. (2005), where the mathematics are translated into the bond-graph formalism. This describes the model in energetic terms and forces adherence to the laws of thermodynamics.
Three modules are presented:

Fig. 1. Overview of modules in this FCU
The bond-graph network for these modules are as follows:
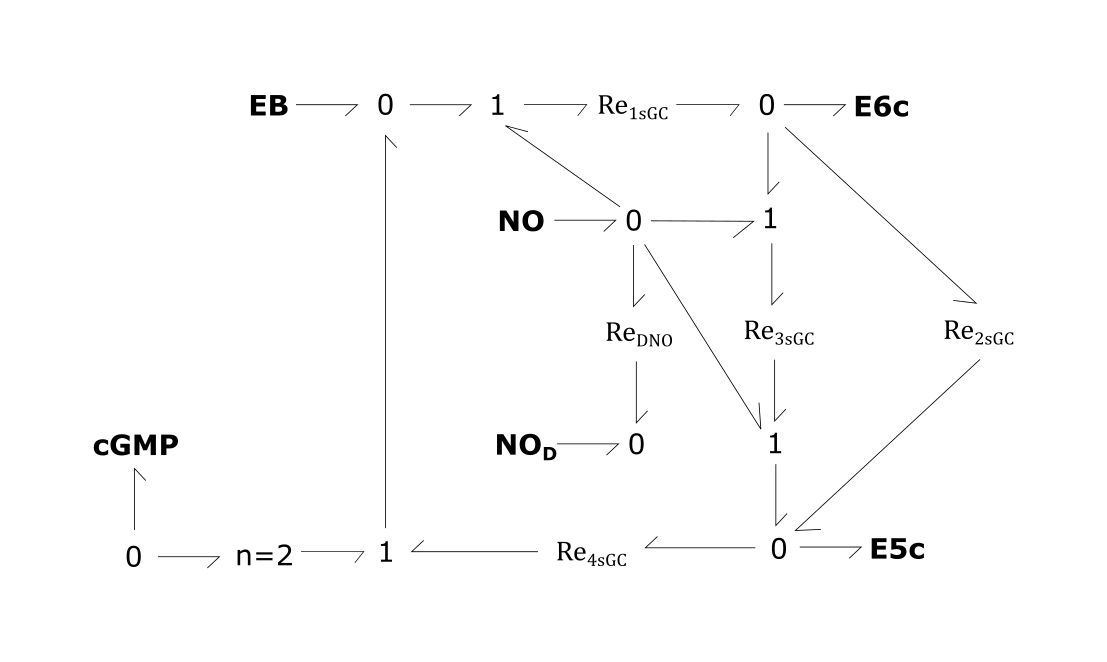
Fig. 2. sGC activation module
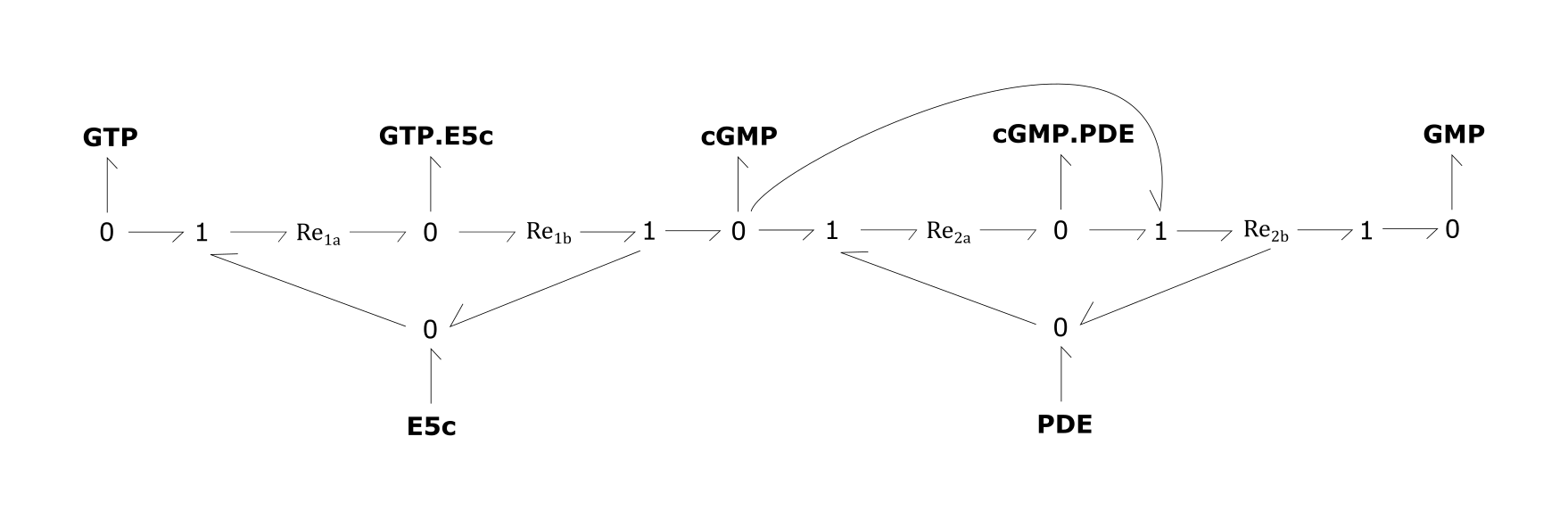
Fig. 3. cGMP production and degradation module
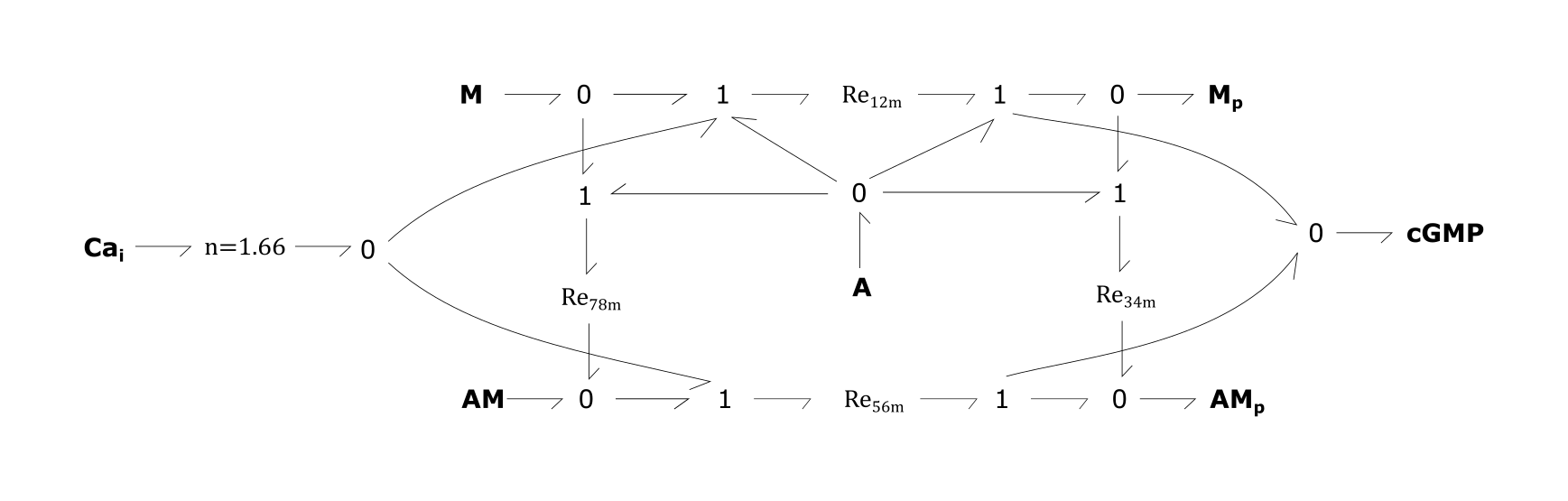
Fig. 4. Smooth muscle contraction module
For the above bond-graphs, a '0' node refers to a junction where all chemical potentials are the same. A '1' node refers to all fluxes being the same going in and out of the junction.
| Abbreviation | Name |
|---|---|
| sGC | Guanylyl cyclase (soluble) |
| EB | Basal variant of sGC |
| E6c | Intermediate variant of sGC |
| E5c | Active variant of sGC |
| cGMP | Cyclic guanosine monophosphate |
| GMP | Guanosine monophosphate |
| GTP | Guanosine triphosphate |
| NO | Nitric oxide |
| NOD | Nitric oxide product (unavailable for this scheme) |
| PDE | Phosphodiesterase-5 |
| M | Myosin light chain |
| Mp | Myosin light chain, phosphorylated |
| A | Actin |
| AM | Latch bridge |
| AMp | Cross bridge |
| Ca | Calcium ion |
Parameter finding
A description of the process to find bond-graph parameters is shown in the folder parameter_finder, which relies on the:
- stoichiometry of system
- kinetic constants for forward/reverse reactions
- If not already, all reactions are made reversible by assigning a small value to the reverse direction.
Here, this solve process is performed in Python.
Original kinetic model
Yang et. al.: Mathematical modeling of the nitric oxide/cGMP pathway in the vascular smooth muscle cell.
