FCU_EC_coupling.cellml
About this Functional Cell Unit
This model is a Functional Cell Unit for excitation-contraction coupling in the cardiac cell.
This is a composite module, composed of several individual modules, and merged together in a modular fashion.
- INPUTS:
- Stimulus current or action potential.
- OUTPUTS:
- Change in number of myosin binding sites on cross-bridges.
Model status
The current CellML implementation compiles in OpenCOR.
Model overview
All components are presented in bond-graph form. Components are made by converting an existing kinetic model and translating into bond-graph form.
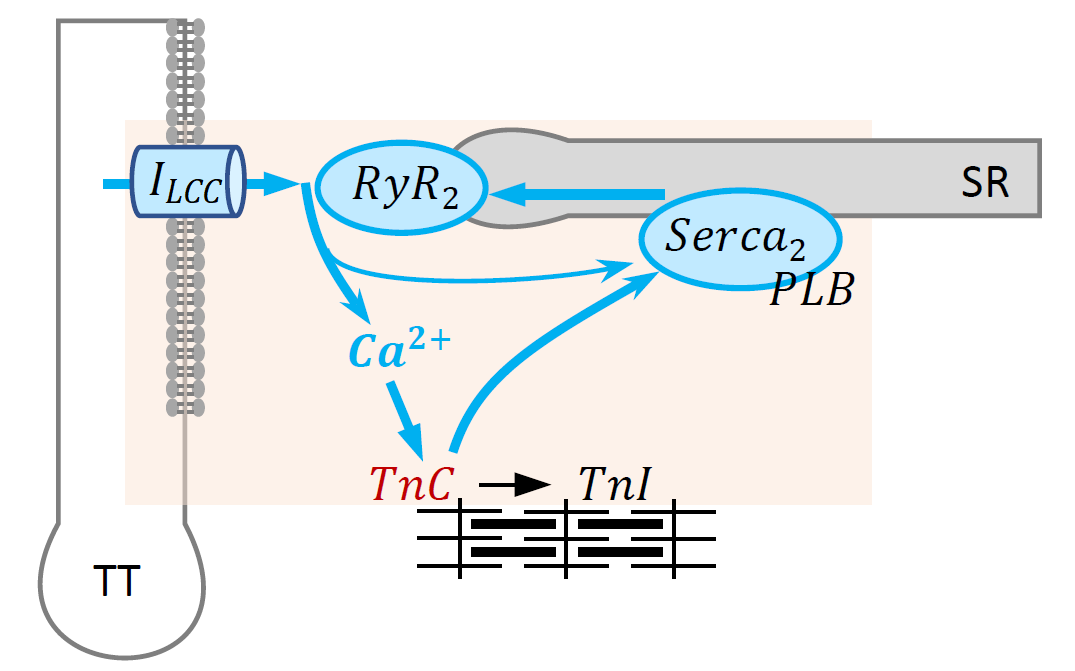
Components of the model.
A description of the process to find bond-graph parameter is shown in the folder parameter_finder, which relies on the:
- stoichiometry of system
- kinetic constants for forward/reverse reactions
- If not already, all reactions are made reversible by assigning a small value to the reverse direction.
- linear algebra script, which outputs the bond-graph parameters K and κ.
Here, this solve process is performed in Python.
Modular description
Components
CellML divides the mathematical model into distinct components, which are able to be re-used in other composite models. These CellML components are:
- Sarcoplasmic/endoplasmic Ca2+ ATPase (SERCA)
- Phospholamban regulation (PLB)
- Ryanodine receptor (RyR)
- L-type calcium channel (LCC)
- Troponin-C (TnC)
| Source | Components |
|---|---|
| Saucerman et al. (2003) | PLB |
| Luo and Rudy (1994) | LCC |
| Tran et al. (2009) | SERCA |
| Stern et al. (1999) | RyR |
| Niederer et al. (2006) | TRPN (TnC) |
Each of these blocks is itself a CellML model, complete with bond-graph parameters appropriate for the isolated system.
